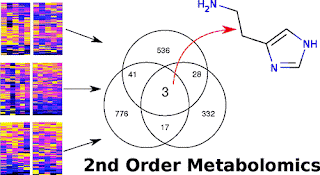
Abstract: Mass spectrometry-based untargeted metabolomics often results in the observation of hundreds to thousands of features that are differentially regulated between sample classes. A major challenge in interpreting the data is distinguishing metabolites that are causally associated with the phenotype of interest from those that are unrelated but altered in downstream pathways as an effect. To facilitate this distinction, here we describe new software called metaXCMS for performing second-order (“meta”) analysis of untargeted metabolomics data from multiple sample groups representing different models of the same phenotype. While the original version of XCMS was designed for the direct comparison of two sample groups, metaXCMS enables meta-analysis of an unlimited number of sample classes to facilitate prioritization of the data and increase the probability of identifying metabolites causally related to the phenotype of interest. metaXCMS is used to import XCMS results that are subsequently filtered, realigned, and ultimately compared to identify shared metabolites that are up- or down-regulated across all sample groups. We demonstrate the software’s utility by identifying histamine as a metabolite that is commonly altered in three different models of pain. metaXCMS is freely available at http://metlin.scripps.edu/metaxcms/.
The full article is available here.
Tuesday, December 21, 2010
Wednesday, December 1, 2010
Hans Rosling shows the best stats you've ever seen
Beautiful visualizations of statistics by Hans Rosling.
Might be inspiring for visualizations of metabolomics time series data ...
There is also the documentary "The Joy of Stats" on BBC in December.
Might be inspiring for visualizations of metabolomics time series data ...
There is also the documentary "The Joy of Stats" on BBC in December.
Tuesday, November 23, 2010
Combined genome-wide association mapping with metabolomics
Interesting article in PLoS Genetics:
The Complex Genetic Architecture of the Metabolome
Association between more than 200,000 single-nucleotide variants across the genome and levels of 327 metabolites in 96 strains of Arabidopsis thaliana showed that only 23–30% of the variation in cellular metabolite levels was associated with specific sites in the genome.
The Complex Genetic Architecture of the Metabolome
Association between more than 200,000 single-nucleotide variants across the genome and levels of 327 metabolites in 96 strains of Arabidopsis thaliana showed that only 23–30% of the variation in cellular metabolite levels was associated with specific sites in the genome.
Monday, November 15, 2010
The Untargeted Metabolomics Workflow
During the past 2 years, the methodology that I have employed to make metabolite identifications using an untargeted metabolomics workflow has evolved. In 2008, not only were metabolite databases smaller, but they also did not have some of the advanced functionality that is available today. For example, searching for sodium and potassium adducts required manually calculating masses from the observed m/z values. We have come a long way with improvements in both metabolomics software and databases facilitating metabolite identification. Major databases emerging as the key players for untargeted studies are HMDB, Lipid Maps, and METLIN. Each, in my opinion, have their own advantages. I would like to start this blog by surveying which databases metabolomics investigators utilize the most frequently and why. HMDB, for example, provides so-called MetaboCards in which fundamental biological facts are introduced for queried molecules. This information can be particularly useful in filtering putative hits for metabolites that may not be relevant to the sample type being analyzed, such as a hit for a plant metabolite from bacterial cell results. Another new function that has been recently incorporated into METLIN is the ability to search fragment ions from MS/MS data. With this function, it is now possible to do MS/MS on all features of interest in a dataset prior to querying databases to potentially reduce false-negative hits. A few years ago, the workflow of identifying metabolites in a global MS-based study offered little room for creativity. I am certain today, however, that investigators are taking advantage of the various new database functionality in a multitude of innovative ways. I hope that by discussing and exchanging ideas about our untargeted workflows we can learn new ways to facilitate what I still would classify as the rate-limiting step in metabolomics, metabolite identification. So what process do you use to make metabolite identifications? How do you prioritize your feature lists? Do you search all the databases on the web, or do you refine yourself to an in-house library? I look forward to reading about your different points of view!
Friday, November 12, 2010
A short history of XCMS
XCMS is an open-source, platform-independent R-package that was developed to perform untargeted metabolite profiling with LC/MS. XCMS reads and processes LC/MS data stored in netcdf , mzXML, mzData and mzML files. It provides method for peak picking, non-linear retention time alignment, visualization, relative quantization and statistics. XCMS is capable of simultaneously preprocessing, analyzing, and visualizing the raw data from hundreds of samples. The original XCMS paper published in 2006 was cited more than 270 times (Google Scholar, 11/12/2010).
Colin Smith initially developed XCMS in 2004. In 2008 Steffen Neumann and Ralf Tautenhahn joined the development team and later in 2009 Paul Benton. Today, XCMS contains two methods for LC/MS feature detection and a method for peak detection in single high-res spectra (FTICR, MALDI, DIMS). Two different non-linear retention time correction methods are available, and two methods to group LC/MS features. A separate method is implemented to align single high-res spectra using a moving-window technique. Mass spectra, TICs, EICs and EIC overlays, 3D LC/MS surface plots and boxplots can be generated by XCMS. Methods to read and preprocess MS/MS data are available. XCMS can make use of multicore processors, as well as MPI or SNOW clusters to speed up the data processing. XCMS is now widely used for untargeted metabolomics, metabolic profiling and biomarker discovery.
Spring 2004: Added methods for reading and displaying raw data from NetCDF files (Colin)
Summer 2004: Developed methods for kernel density peak grouping and LOESS retention time alignment (Colin)
Fall 2004: Developed matched filter peak picker, EIC generation (Colin)
March 2005: Checked into Bioconductor SVN repository (Colin)
December 2005: mzXML, mzData import added (Colin)
April 2007: centWave peak detection added (Ralf)
November 2007: Reading of MS/MS spectra added (Steffen)
January 2008: single spectra alignment method added (Steffen)
July 2008: Multiprocessor peak picking added via MPI (Ralf)
March 2009: OBI-Warp retention time alignment added (Steffen, Ralf)
April 2009: group nearest alignment method added (Steffen, Ralf)
June 2010: gap filler/stitch method added (Paul)
September 2010: 64 bit support added (Steffen, Ralf)
Colin Smith initially developed XCMS in 2004. In 2008 Steffen Neumann and Ralf Tautenhahn joined the development team and later in 2009 Paul Benton. Today, XCMS contains two methods for LC/MS feature detection and a method for peak detection in single high-res spectra (FTICR, MALDI, DIMS). Two different non-linear retention time correction methods are available, and two methods to group LC/MS features. A separate method is implemented to align single high-res spectra using a moving-window technique. Mass spectra, TICs, EICs and EIC overlays, 3D LC/MS surface plots and boxplots can be generated by XCMS. Methods to read and preprocess MS/MS data are available. XCMS can make use of multicore processors, as well as MPI or SNOW clusters to speed up the data processing. XCMS is now widely used for untargeted metabolomics, metabolic profiling and biomarker discovery.
Spring 2004: Added methods for reading and displaying raw data from NetCDF files (Colin)
Summer 2004: Developed methods for kernel density peak grouping and LOESS retention time alignment (Colin)
Fall 2004: Developed matched filter peak picker, EIC generation (Colin)
March 2005: Checked into Bioconductor SVN repository (Colin)
December 2005: mzXML, mzData import added (Colin)
April 2007: centWave peak detection added (Ralf)
November 2007: Reading of MS/MS spectra added (Steffen)
January 2008: single spectra alignment method added (Steffen)
July 2008: Multiprocessor peak picking added via MPI (Ralf)
March 2009: OBI-Warp retention time alignment added (Steffen, Ralf)
April 2009: group nearest alignment method added (Steffen, Ralf)
June 2010: gap filler/stitch method added (Paul)
September 2010: 64 bit support added (Steffen, Ralf)
Friday, October 29, 2010
What is a feature in metabolomics?
After four years working on numerous projects using untargeted metabolomics, called by some colleagues "global metabolite profiling", my vision of this scientific discipline has been evolving largely as a result of the difficulties I've been encountering. Starting with the sample preparation (i.e., extraction of metabolites), through data processing, and ending with the identification of metabolites, each of these steps has caused me its own headaches. Still, I must admit, I have greatly simplified the whole process and now I can conduct much more pragmatic metabolomics studies. From this blog, I would like to begin a series of dialogues aimed at discussing the various methodological aspects of metabolomics. And I want to start with, perhaps, the subject that has suffered the largest transformation in my methodological workflow: data processing. Those who work with TOF instruments coupled to liquid chromatography will be familiar with the massive amount of data obtained with programs like XCMS. Well, in my opinion it all comes down to understanding the term “feature”. A very simple definition of feature is “a molecular entity with a unique m/z and retention time”. However, one feature does not necessarily correspond to a metabolite. The number of features is always much higher than the number of metabolites. How much? How many features do you typically detect in a regular untargeted metabolomics study? How do you filter features to end up identifying metabolites?
 |
| feature detection with XCMS |
These are some of the points I would like to discuss here. I bet you'll read a lot of different opinions on this matter, and I hope you can convey my experience and I can learn a bit more of all your points of view.
Welcome to metaBlogOmics!
Oscar
Subscribe to:
Posts (Atom)