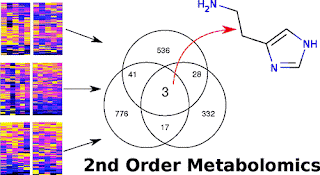
Abstract: Mass spectrometry-based untargeted metabolomics often results in the observation of hundreds to thousands of features that are differentially regulated between sample classes. A major challenge in interpreting the data is distinguishing metabolites that are causally associated with the phenotype of interest from those that are unrelated but altered in downstream pathways as an effect. To facilitate this distinction, here we describe new software called metaXCMS for performing second-order (“meta”) analysis of untargeted metabolomics data from multiple sample groups representing different models of the same phenotype. While the original version of XCMS was designed for the direct comparison of two sample groups, metaXCMS enables meta-analysis of an unlimited number of sample classes to facilitate prioritization of the data and increase the probability of identifying metabolites causally related to the phenotype of interest. metaXCMS is used to import XCMS results that are subsequently filtered, realigned, and ultimately compared to identify shared metabolites that are up- or down-regulated across all sample groups. We demonstrate the software’s utility by identifying histamine as a metabolite that is commonly altered in three different models of pain. metaXCMS is freely available at http://metlin.scripps.edu/metaxcms/.
The full article is available here.
Tuesday, December 21, 2010
Wednesday, December 1, 2010
Hans Rosling shows the best stats you've ever seen
Beautiful visualizations of statistics by Hans Rosling.
Might be inspiring for visualizations of metabolomics time series data ...
There is also the documentary "The Joy of Stats" on BBC in December.
Might be inspiring for visualizations of metabolomics time series data ...
There is also the documentary "The Joy of Stats" on BBC in December.
Subscribe to:
Comments (Atom)